Is a stool sample sufficient for diagnosing Crohn's disease?
An international research group led by Victoria Pascal from the Department of Gastroenterology, Vall d'Hebron Research Institute in Barcelona, has succeeded in identifying patients with Crohn's disease (CD) solely from stool samples.
The method is independent of geographical particularities
An international research group led by Victoria Pascal from the Department of Gastroenterology, Vall d'Hebron Research Institute in Barcelona, has succeeded in identifying patients with Crohn's disease (CD) solely from stool samples.
With a sensitivity of 80% and a specificity of 89 to 94%, scientists were able to distinguish Crohn patients from healthy control patients, patients with ulcerative colitis (UC), irritable bowel syndrome (IBS) and anorexia nervosa (AN). The following proved to be diagnostically effective in:
- reduced diversity,
- the overall reduced stability of the intestinal flora of Crohn's patients, and
- a group of eight intestinal germs whose distribution was characteristically altered in Crohn's disease.
The authors assume that their non-invasive method can enable the diagnosis of Crohn's disease if no specific characteristics of the disease can or have been detected in its early phase. Due to the equally high reliability of stool samples from four European countries, the researchers assume that their method is independent of geographical characteristics of the intestinal flora.
Previous microbiomics studies have been contradictory
The starting point of the study was the search for a clear dysbiosis assignment to chronic inflammatory bowel diseases (IBD), Crohn's disease (CD), and ulcerative colitis (UC). The scientists first analyzed stool samples from 178 subjects from Spain (including healthy, unrelated control subjects [HC]: 40, patients with CD: 34; patients with UC: 33; healthy related control subjects [HR]: 36 for CD patients, 35 for UC patients). Subsequently, the study was repeated on a comparable population in Belgium and subsequently extended to a total of 2,045 participants from Spain, Belgium, Great Britain and Germany (HC: 1247, CD: 339, UC: 158, IBS: 202, AN: 99).
To distinguish the microbial genera present in the samples, the researchers used 16S rRNA sequencing of ribosomal RNA. The statistical analysis of the 115 million amino acid sequences obtained was used by the UniFrac metric to express the differences between the encountered intestinal flora in the form of a biological distance (see Fig. 1C) (see Source 2).
Crohn patients can be identified by their intestinal flora
The evaluation of the stool samples showed the following results (see Fig. 1):
- Based on the frequency distribution of eight genera in the intestinal flora, a method was developed to identify patients with Crohn's disease and to distinguish them from patients with UC (see Fig.1).
- Healthy related and unrelated controls had comparable microbiomes except for the genus Collinsiella, which increased among relatives of IBD patients.
- Patients with CD and UC differed significantly from both control groups in terms of their microbiome (p = 0.001).
- CD patients had a higher intestinal flora instability (p < 0.001) compared to UC patients.
- CD patients, but not UC patients, showed a reduced alpha diversity of their microbioma compared to both control groups.
- No significant change in intestinal flora was observed in CD and UC patients compared to patients who remained in remission.
- No biomarkers were identified that indicated an impending outbreak of the disease.
- The microbiome in CD patients seems to be more characterized by a loss of useful bacteria (Faecalibacterium, Christensenellaceae, Methanobrevibacter, Oscillospira) than by an increase of pathogenic germs (Fusobacterium, Escherichia).
- A connection between the microbiome and the current or previous smoking behavior of IBD patients could not be proven.
- Fecal calprotectin was significantly elevated in IBD patients compared to healthy controls but was not suitable for differentiating between UC and CD.
- Streptococci in the microbiome proved to be a marker for an increased risk of postoperative relapse in CD patients.
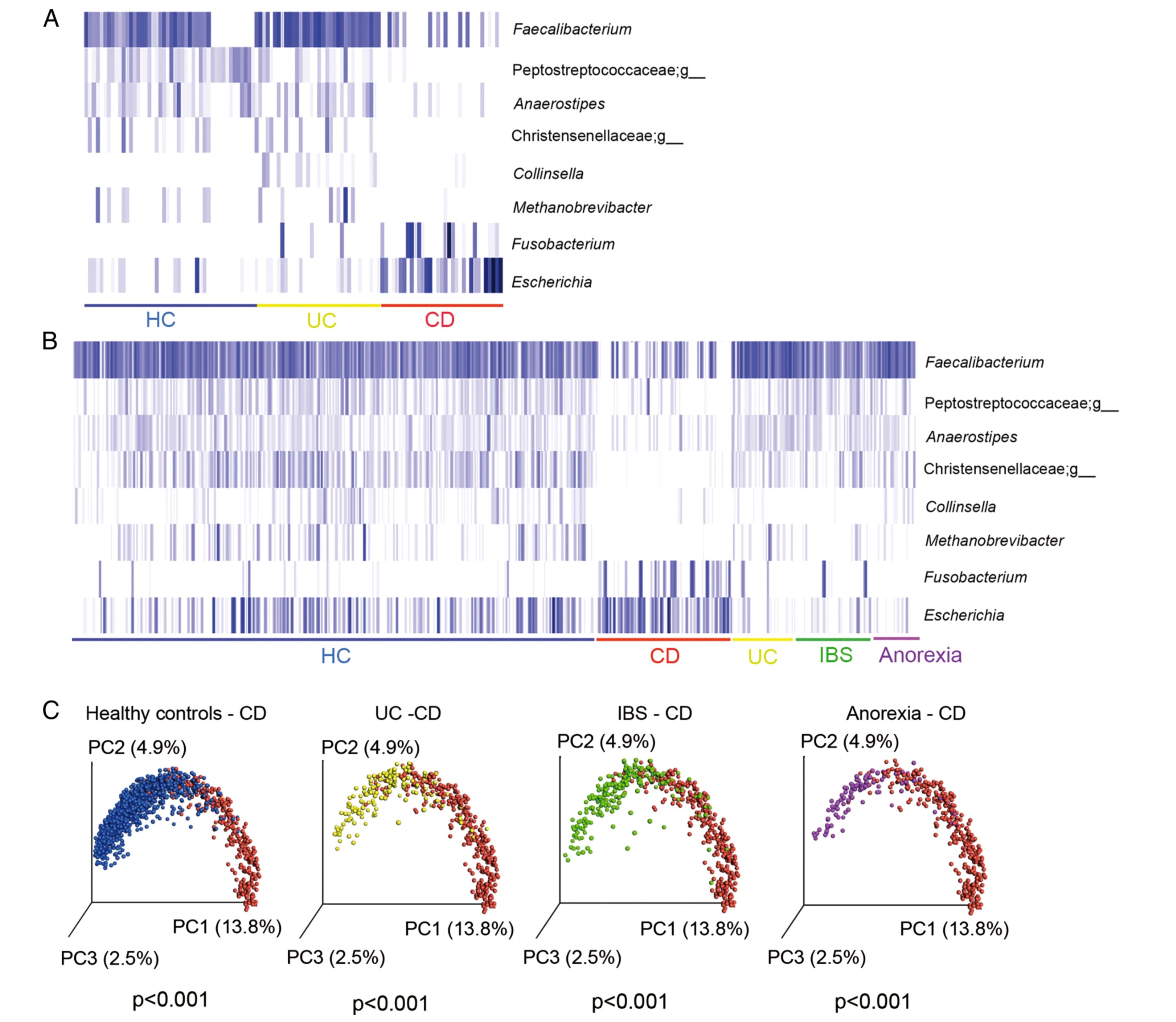
Fig. 1: Microbial markers to distinguish CD and UC (A) in the discovery study (n = 178), (B) in the verification study (n = 2,045). Blue strokes indicate proof of the corresponding genus, whereby lighter shades indicate lower and darker shades indicate greater colonization. Patient groups are marked by colour: blue: HC and HR; red: CD; yellow: UC; green: IBS; violet: AN. (C) UniFrac Principal Coordinate Analysis with the coordinates PC1, PC2, PC3 between two patient groups. In all four partial illustrations, patients with Crohn's disease (CD) proved to be significantly different from the respective comparison group (p < 0.001) (Sources: 1)
Sources:
1. Pascal V et al. A microbial signature for Crohn's disease. Good 2017;66:813-822.
2nd Lozupone C, Knight R. UniFrac: A New Phylogenetic Method for Comparing Microbial Communities. Applied and Environmental Microbiology 2005:71(12):8228-8235.