New software predicts the fate of individual cells
"scVelo" is a machine learning-based open software that predicts gene activity in individual cells. This could allow predicting the future state of individual cells and to better understand the course of a disease.
The technology helps to better understand the course of a disease
The study of cell dynamics provides a deeper insight into the formation and development of cells and a better understanding of the course of a disease. A research team at the Munich-based Helmholtz Zentrum and Munich Technical University (German acronym: TUM) have developed "scVelo", a method based on machine learning and open-source software that can predict the dynamics of gene activity in individual cells. This enables researchers to predict the future state of individual cells.
Conventional methods for single-cell sequencing make it possible to gain insights into differences and functions at the cellular level - but this can only be done as a static snapshot and not as a timelapse. This limitation makes it difficult to draw conclusions about cell dynamics. With the recently introduced "RNA Velocity" method, the developmental course of a single cell can be reconstructed mathematically (based on the ratio of spliced and unspliced RNA transcripts). However, this method has so far only been applicable to static cell populations. The researchers, therefore, looked for ways to methodically extend the concept of "RNA velocity" so that it could also be applied to dynamic populations. Such cell populations are of crucial importance for understanding cell development and reactions to diseases.
"Single-cell velocity"
A team from the Institute of Computational Biology at Helmholtz Zentrum München and the Faculty of Mathematics at TUM developed "scVelo" (single-cell velocity). The method determines the RNA velocity using a model based on artificial intelligence (AI). The AI learns the entire transcription dynamics for each gene. This enables researchers to apply the concept of RNA velocity to a variety of biological systems, including dynamic populations.
"We used scVelo to decipher cell development in the endocrine pancreas and hippocampus. We have also investigated dynamic processes during lung regeneration - and this is just the beginning," says Volker Bergen, developer of scVelo and main author of the related publication in Nature Biotechnology.
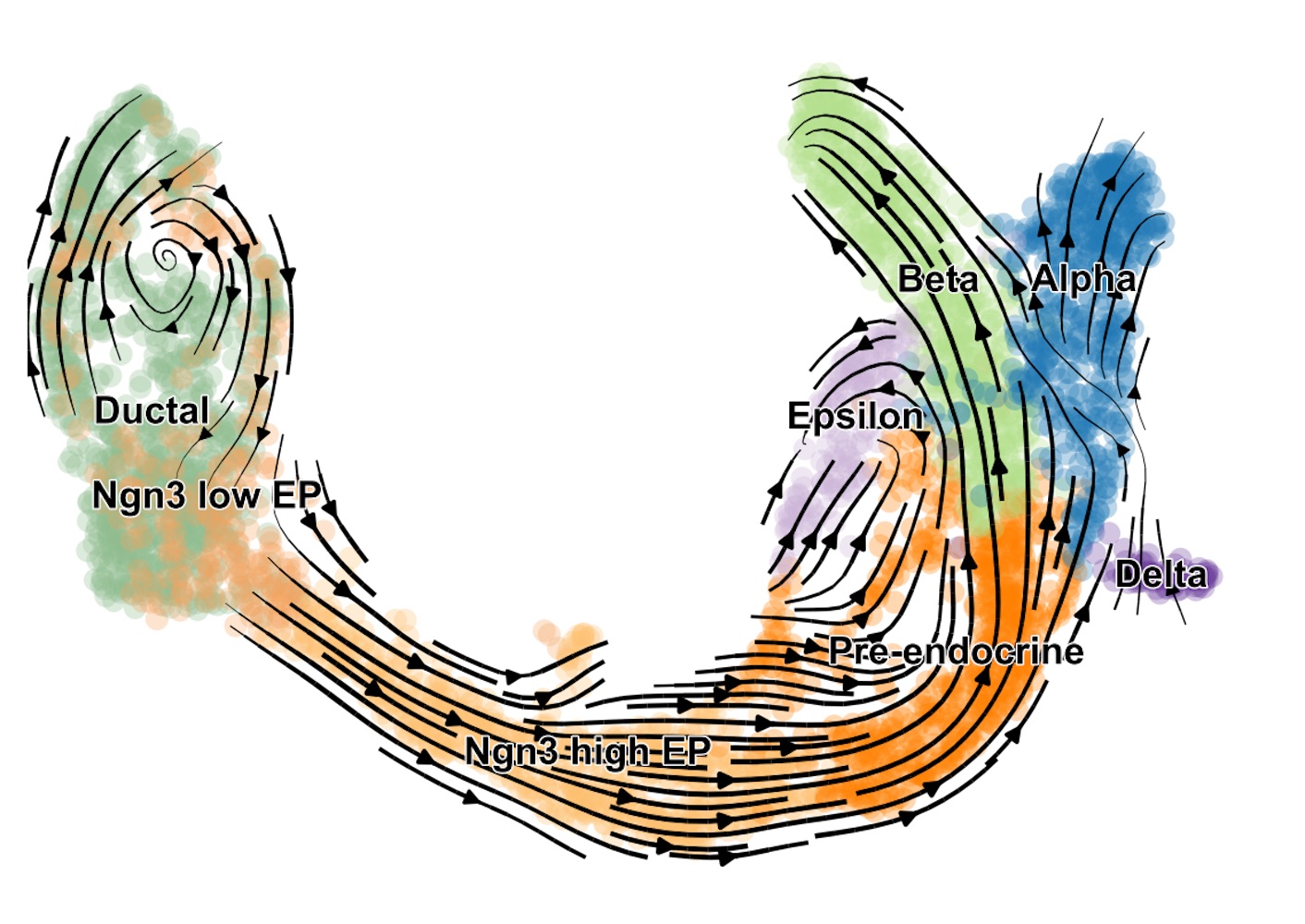
scVelo provides a detailed insight into the developmental processes of the pancreas.
Image credit: Helmholtz Zentrum München
With scVelo, researchers can determine reaction rates without the need for time-consuming experiments and thus find out the speed at which RNA is produced and spliced and how quickly it finally decays. These rates can help to better understand cell identity and phenotypic differences. By introducing latency, they can completely reconstruct cell development and position each cell along its developmental path. In particular, this makes the decision-making processes of a cell easier to understand. In addition, scVelo uncovers regulatory changes and identifies the genes that might be responsible for these changes. This enables researchers to understand not only how, but also why cells develop in a certain way.
Personalized treatment methods
AI-based solutions such as scVelo could help in the development of personalized treatment methods. The leap from static snapshots to fully dynamic systems allows researchers to move from purely descriptive to predictive models. In the future, this could help to better understand disease progression, for example, tumor formation, or decipher the cell response in response to cancer treatment.
"scVelo has been downloaded almost 60,000 times since its publication last year. The software has become a popular and important tool for the development of kinetic models for single-cell transcriptomics," adds Prof. Fabian Theis, who designed the study and is Director at the Institute of Computational Biology at the Munich Helmholtz Zentrum and Chair of Mathematical Modeling of Biological Systems at TUM.
Source:
Bergen et al., 2020: Generalizing RNA velocity to transient cell states through dynamical modeling